Tech
Supercomputing in the AI era accelerates protein structure prediction
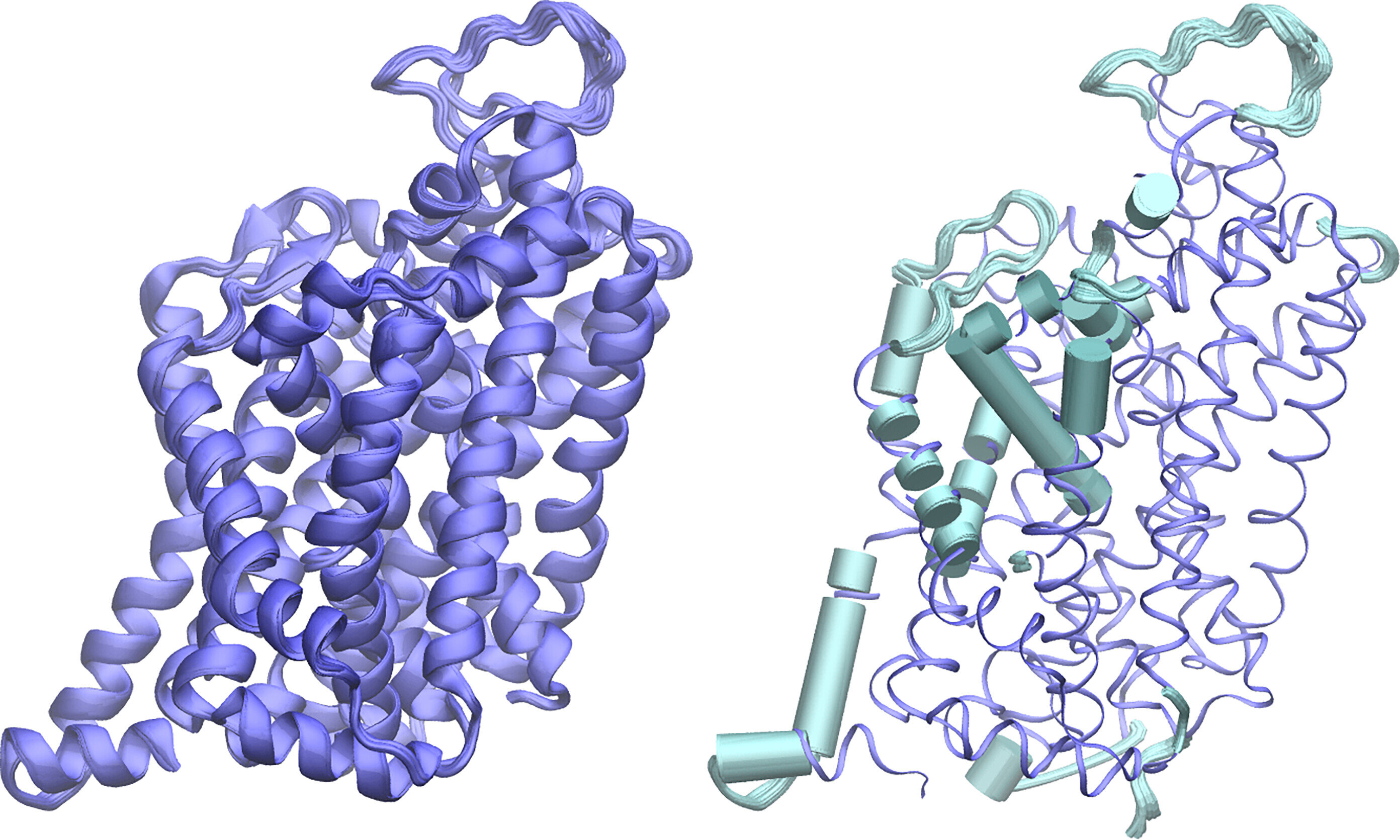
Protein structure used to test APACE: serotonin transporter (PDB accession number: 6AWO, abbreviated SERT). The left panel shows an ensemble of 100 SERT predicted conformations overlaid with the actual SERT, which matches well. The right panel shows the highly mutated transmembrane domain (shown in cyan) overlaid with root-mean-square fluctuations. Credit: Proceedings of the National Academy of Sciences (2024). DOI: 10.1073/pnas.2311888121
For researchers, using high-performance computers can be a bit intimidating: Understanding the best interfaces, how to extend the software, and working with huge data sets require unique expertise.
Fortunately, NCSA does more than just deploy and operate these powerful systems: the center is also home to a Science and Engineering Applications Support (SEAS) team dedicated to helping researchers make efficient use of the hardware and software resources available at NCSA.
By working with SEAS, researchers can get support with installing Python packages, learn how to choose the best parallel computing engine for their projects, and successfully deploy artificial intelligence models thanks to a groundbreaking study published in the journal PNASlearn, titled “APACE: AlphaFold2 and Advanced Computing as a Service to Accelerate Discovery in Biophysics.”
The PNAS research paper, authored by Roland Haas, senior research programmer in the SEAS group, Eliu Huerta, translational AI lead at the U.S. Department of Energy (DOE) Argonne National Laboratory and CASE senior scientist at the University of Chicago, Hyun Park, then a biophysics doctoral student at the University of Illinois, and Parth Patel, a graduate research assistant at NCSA, describes a new computational framework that simplifies and speeds up the process of using AI tools and algorithms to understand three-dimensional protein structures.
The framework also predicts the structural diversity of proteins, an important property because proteins are flexible structures that can switch between different conformations to fulfill their functions.
The research team developed APACE, a computational tool that effectively handles AlphaFold2, an AI program used to predict protein structures on high-performance computing systems. The team deployed APACE on NCSA's Delta supercomputer and measured its performance in predicting the structures of four representative proteins.
Using an ensemble of up to 300 distributed across 300 NVIDIA A100 GPUs, we find that APACE is up to two orders of magnitude faster than an off-the-shelf AlphaFold2 implementation.
Moreover, the same approach can be used across a range of scientific disciplines and can even be used in conjunction with robotics laboratories to automate and accelerate scientific discovery. The team then replicated this work on the Polaris supercomputer at the Argonne Leadership Computing Facility, a Department of Energy Office of Science User Facility.
“The underlying AI models have the potential to transform the practice of science if they are searchable, accessible, and ready to be used by the broader scientific community,” said Huerta. “This project shows how we can truly democratize cutting-edge AI and leverage modern computing environments to create and share the scientific data infrastructure needed to maximize the reach of science.”
Biomedical researchers study proteins to understand a wide range of biological functions. Proteins are chains of amino acids whose 3D structural arrangement determines their biological function.
Understanding how proteins are formed – where amino acids assemble into structured chains that can perform specific functions, in a process called protein folding – is important not only for understanding normal biological function but also for understanding why misfolding can lead to serious diseases.
Predicting protein folds is extremely computationally intensive, as a typical protein can contain hundreds of amino acids and thousands of molecules that may bind in different ways.
The usual methods for studying protein structure are X-ray crystallography, a tool that determines the atomic and molecular structure of crystals, and cryo-electron microscopy, in which molecules are flash-frozen in liquid nitrogen, bombarded with electrons and images taken with a special camera.
AlphaFold and AlphaFold2 have demonstrated that AI software can accurately and rapidly predict protein structures from amino acid sequences, and the development of APACE builds on this breakthrough.
APACE has optimized AlphaFold2 to run at scale on high-performance computing platforms, effectively handling multi-terabyte protein databases. This work shows that combining large-scale AI models with the power of high-performance computing enables scientists to study multiple protein complexes and obtain results quickly, accurately, and at high resolution. All of this could lead to a more complete understanding of protein structure and facilitate the development of new drugs that can treat many diseases.
“Research into new drugs is extremely time-consuming and is bottlenecked by the need to synthesize various candidate compounds in the laboratory to test their medical effects,” Haas said.
APACE allows drug researchers to significantly reduce the time needed to screen potential candidate compounds and focus on the most promising ones, allowing more compounds to be tested and shortening the development time for new drugs, such as those tailored to specific virus strains.
A key feature of APACE is improved data management, achieved by hosting AlphaFold2's multi-terabyte models and databases on supercomputers, from which the framework's neural networks can easily access the data. Other improvements include CPU and GPU optimizations, parallelizing the GPU-intensive protein structure prediction step of the neural network.
“The first problem with using an AI model is storing the data,” said Park, who, like Patel, was an intern at Argonne National Laboratory when the APACE research was being conducted.
“In addition to the computations from sequence to structure prediction, you need to hand over 2.6 terabytes (the size of the AlphaFold2 database). Some university labs might be able to do that, but the key is to scale it up so that it can be used by scientists all over the world.”
Patel added, “That's why using HPC is so important, especially for AI models. Anyone with access to an HPC system has access to both the data and the computing power to run the actual AI model calculations, not to mention the massive speed improvements.”
Huerta said the team chose to use AlphaFold2 because it is widely used across different research communities, including biophysics, chemistry, and drug design and discovery.
“APACE provides all the capabilities of the original AlphaFold2 model, enabling researchers to leverage supercomputers to accelerate time to solution and connect this tool to self-driving labs to automate and accelerate discovery,” he said.
Huerta said the team will continue to build a community of APACE users to maximize the usability of AI models on HPC platforms. Haas said the team is now focused on eliminating remaining bottlenecks in the system to further improve speed. They also want to make APACE available on more computing clusters so more scientists can take advantage of it.
“We also want to explore using the techniques we developed to accelerate Alphafold2 on other fundamental machine learning models that are too complex to be easily used on a typical desktop workstation,” Haas says. “It's all about making the best tools as easy to use as possible.”
More information: Hyun Park et al., “APACE: AlphaFold2 and Advanced Computing as a Service to Accelerate Discovery in Biophysics,” Proceedings of the National Academy of Sciences of the United States of America (2024). DOI: 10.1073/pnas.2311888121
Provided by National Supercomputing Applications Center
Citation: Supercomputing in the AI era accelerates protein structure prediction (June 28, 2024) Retrieved June 28, 2024 from https://phys.org/news/2024-06-supercomputing-age-ai-protein.html
This document is subject to copyright. It may not be reproduced without written permission, except for fair dealing for the purposes of personal study or research. The content is provided for informational purposes only.
|
Sources 2/ https://phys.org/news/2024-06-supercomputing-age-ai-protein.html The mention sources can contact us to remove/changing this article |
What Are The Main Benefits Of Comparing Car Insurance Quotes Online
LOS ANGELES, CA / ACCESSWIRE / June 24, 2020, / Compare-autoinsurance.Org has launched a new blog post that presents the main benefits of comparing multiple car insurance quotes. For more info and free online quotes, please visit https://compare-autoinsurance.Org/the-advantages-of-comparing-prices-with-car-insurance-quotes-online/ The modern society has numerous technological advantages. One important advantage is the speed at which information is sent and received. With the help of the internet, the shopping habits of many persons have drastically changed. The car insurance industry hasn't remained untouched by these changes. On the internet, drivers can compare insurance prices and find out which sellers have the best offers. View photos The advantages of comparing online car insurance quotes are the following: Online quotes can be obtained from anywhere and at any time. Unlike physical insurance agencies, websites don't have a specific schedule and they are available at any time. Drivers that have busy working schedules, can compare quotes from anywhere and at any time, even at midnight. Multiple choices. Almost all insurance providers, no matter if they are well-known brands or just local insurers, have an online presence. Online quotes will allow policyholders the chance to discover multiple insurance companies and check their prices. Drivers are no longer required to get quotes from just a few known insurance companies. Also, local and regional insurers can provide lower insurance rates for the same services. Accurate insurance estimates. Online quotes can only be accurate if the customers provide accurate and real info about their car models and driving history. Lying about past driving incidents can make the price estimates to be lower, but when dealing with an insurance company lying to them is useless. Usually, insurance companies will do research about a potential customer before granting him coverage. Online quotes can be sorted easily. Although drivers are recommended to not choose a policy just based on its price, drivers can easily sort quotes by insurance price. Using brokerage websites will allow drivers to get quotes from multiple insurers, thus making the comparison faster and easier. For additional info, money-saving tips, and free car insurance quotes, visit https://compare-autoinsurance.Org/ Compare-autoinsurance.Org is an online provider of life, home, health, and auto insurance quotes. This website is unique because it does not simply stick to one kind of insurance provider, but brings the clients the best deals from many different online insurance carriers. In this way, clients have access to offers from multiple carriers all in one place: this website. On this site, customers have access to quotes for insurance plans from various agencies, such as local or nationwide agencies, brand names insurance companies, etc. "Online quotes can easily help drivers obtain better car insurance deals. All they have to do is to complete an online form with accurate and real info, then compare prices", said Russell Rabichev, Marketing Director of Internet Marketing Company. CONTACT: Company Name: Internet Marketing CompanyPerson for contact Name: Gurgu CPhone Number: (818) 359-3898Email: [email protected]: https://compare-autoinsurance.Org/ SOURCE: Compare-autoinsurance.Org View source version on accesswire.Com:https://www.Accesswire.Com/595055/What-Are-The-Main-Benefits-Of-Comparing-Car-Insurance-Quotes-Online View photos
to request, modification Contact us at Here or [email protected]



